Nuisance signal regression
(i) estimate the presence of nuisance signal in the brain,
(ii) control for nuisance signal in task activation GLM analyses, and
(iii) remove nuisance signal as a preprocessing step in preparing data for functional connectivity analyses.
QuNex can handle several types of nuisance signals, which can be divided into three groups:
(i) motion correction parameters,
(ii) nuisance signal extracted from BOLD time series, and
(iii) any externally recorded nuisance signal(s).
We'll review how each type of spurious signal is prepared and regressed.
Regression of movement correction parameters
Head movement during BOLD signal acquisition is an important source of nuisance signal that can mask or inflate the task-related BOLD response and cause spurious correlations in analyses of functional connectivity. Removal of signals related to movement correction parameters is a standard step in
functional connectivity preprocessing and is sometimes also used in task activation analyses. QuNex provides specific functionality for motion correction parameter regression that includes (a) movement parameter preparation and (b) specification of movement parameter regression in GLM modeling.
Motion parameters are automatically processed and copied when data are imported from a minimal preprocessing pipeline into the QuNex folder structure. When data is processed using HCP minimal processing pipelines and imported into the QuNex folder structure using the map_hcp_data command, the six motion parameters (translation in the x, y, and z directions and rotation about the X, Y, and Z axes) are copied to the sessions/<session_id>/images/functional/movement folder. For each bold file, a bold[N]_mov.dat text file is created with the following information:
#frame dx(mm) dy(mm) dz(mm) X(deg) Y(deg) Z(deg)
1 -0.003906 -0.007090 0.013321 0.525369 0.722412 0.611740
2 -0.004437 -0.006768 0.013317 0.523251 0.702507 0.632201
3 -0.005018 -0.007351 0.013436 0.526239 0.684650 0.647907
4 -0.003611 -0.007624 0.013978 0.518844 0.694919 0.785924
5 -0.002795 -0.007855 0.013742 0.497878 0.653377 0.806707
...
The file specifies the translation and rotation of each frame relative to the previous frame (or a reference image in the case of the first frame).
This data is automatically associated with the corresponding fMRI image by the recorded number of the bold image in the file name. For example, bold3_mov.data stores the motion correction parameters for the third bold image as listed in the batch.txt file and as also named in the functional folder (e.g., bold3.nii.gz).
The motion correction parameters are then regressed during GLM modeling, implemented by preprocess_bold and preprocess_conc commands. The regression of the motion parameters is specified with the --bold_nuisance parameter. Specifically, QuNex supports the following options for motion
regression, which can be specified in a comma-separated string:
m– include the six motion correction parameters in the regression,m1d– include the first derivative of the six motion correction parameters in the regression,mSq– include the squared six motion correction parameters in the regression,m1dSq– include the first derivative of the squared six motion correction parameters in the regression.
The motion parameters are included in the GLM as indicated. Estimates for each of the specified regressors are available in the coefficient file, and their estimated contribution to the BOLD signal is removed from the residual time series.
Regression of nuisance signal extracted from BOLD time series
The nuisance signal can be estimated and extracted from the BOLD time series itself. Three types of nuisance signal estimates are most commonly extracted and used: (a) ventricular signal (V), (b) white matter signal (WM), and (c) whole-brain signal (WB). The reasoning is as follows. For the V and WM signals, we assume that the cerebrospinal fluid and white matter, respectively, do not contain information about the neuronal activity. Any changes in the BOLD signal observed in this tissue, therefore, do not reflect neuronal activity, but rather random MR noise, motion artifacts, and/or the effects of physiological changes unrelated to neuronal activity (WM).
The rationale for removing the global signal is a little different. It assumes that although the activity of individual brain regions changes over time, the average activity throughout the brain remains relatively stable, so that any signal present throughout the brain should primarily represent a nonneuronal nuisance signal. This reasoning is still controversial. Research has shown that regression of the global signal can shift the mean of the signal and inflate the negative correlation between brain regions, and that the global signal itself may contain information of interest. Therefore, the decision whether or not to remove the global signal should be made by the researcher on an informed basis, taking into account the potential impact and the specific research question.
Extraction of ventricle, white matter and whole-brain signal
Similar to the motion correction parameters, QuNex provides specific functionality for extracting and regressing BOLD time series based nuisance signal. The extraction of BOLD nuisance signal is supported by the extract_nuisance_signal command. The extract_nuisance_signal command works with a batch.txt file to loop through all sessions and functional images and extract the nuisance signal from each one, taking into account any filtering of sessions and/or images specified by the user. The command always works with volume representations of functional images, because - compared to dense or parcelled CIFTI files - only these images contain the relevant information.
By default, extract_nuisance_signal performs the following steps: First, it loads the volume representation of a functional run (e.g., bold3.nii.gz) stored in the sessions/<session_id>/functional folder. Second, it loads a BOLD brain mask specific to that functional image, located in the sessions/<session_id>/images/segmentation/boldmasks/ folder, e.g., bold3_frame1_brain_mask.nii.gz, and masks out all non-brain voxels so that they are not included in the signal extraction. Third, it loads the freesurfer brain segmentation image for that session, downsampled to functional image resolution, located in sessions/<session_id>/images/segmentation/freesurfer/mri/aparc+aseg_bold.nii.gz.
Based on the segmentation, three masks are defined: a ventricle mask (V) containing all voxels identified as any of the ventricles, a white matter mask (WM) containing all voxels identified as white matter, and a whole-brain mask (WB) that contains all voxels identified as gray matter. After these masks are created, it erodes them by removing all voxels that share a surface (for V and WM masks) or more than nine edges (for WB mask) with voxels not included in the mask. Then it computes and saves the mean signal across all the voxels in each of the masks.
The following files are saved when running extract_nuisance_signal command:
sessions/<session_id>/images/functional/motion/bold[N].nuisance
stores mean signal values across the nuisance ROI for each of the frames in the BOLD imagesessions/<session_id>/images/ROI/nuisance/bold[N]_nuisance.nii.gz
stores a volume image with the following frames:- frame 1: the first volume of the bold image
- frame 2: the whole-brain (WB) mask
- frame 3: the ventricle (V) mask
- frame 4: the white matter (WM) mask
- frame 6: a combined mask with values 1–WB, 2—V, 3–WM
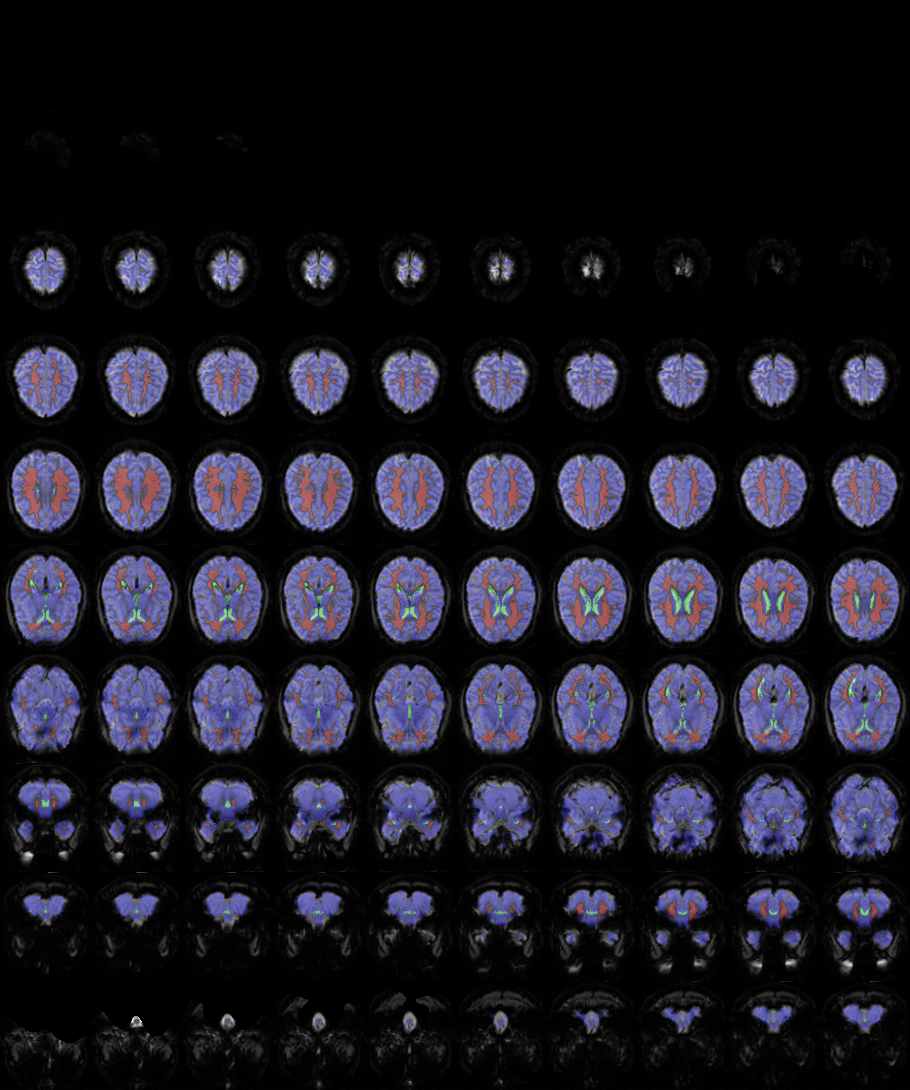
sessions/<session_id>/images/ROI/nuisance//bold1_nuisance.png
an image file with axial slices of the BOLD image of the brain with transparent ovelay indicating whole-brain (blue), ventricle (green) and white matter (red) tissue as used for extracting the nuisance signal
An example .nuisance file is:
frame V WM WB
1 33320.7304687500 26928.9746093750 29940.0703125000
2 33382.0859375000 26915.7148437500 29923.3105468750
3 33334.6640625000 26930.1347656250 29930.4296875000
4 33267.6640625000 26920.2167968750 29926.7617187500
5 33280.3476562500 26907.8964843750 29906.0507812500
...
An example image file of V, WM and WB nuisance masks.
Advance extraction of nuisance BOLD signal
The extract_nuisance_signal provides additional advanced functionality:
Switching off the mask erosion.
Mask erosion is used to remove all boundary voxels between the three tissues of interest, since boundary voxels may contain a mixture of signals due to low resolution and spatial smoothing performed during minimal preprocessing. If desired, this erosion can be turned off by setting the --shrinknsroi parameter to false.
Exclusion of brain areas from the whole-brain mask.
A researcher may wish to exclude certain brain areas from the calculation of the whole-brain signal. For example, if a researcher is interested in functional connectivity between a set of brain regions or in computing a seed map of functional connectivity with a particular region, they may wish to exclude that region(s) from the calculation of the global signal. This is enabled by the --wbmask parameter, which can be either a .names ROI definition file or a binary mask that identifies all voxels that should not be included in the whole-brain nuisance signal mask.
Inclusion of additional regions for the nuisance signal extraction.
At times, a researcher may wish to extract nuisance signals from one or more additional regions inside or outside the brain. This can be accomplished with the parameter --nroi. The parameter can specify either a binary mask file or a .names ROI definition file. In the latter case, the regions to be included must also be specified as a comma-separated list following the path to the .names file, with the two pieces of information separated by a pipe <path to the .names file>|<roi 1>,<roi 2>,<roi 3>. A concrete example could be <path>/nuisance_roi.names|eyes,scull. The .names file can refer to subject-specific masks, which must then be specified with the --sessionroi parameter. The parameter currently accepts 'wb' or 'aseg' as valid values referring to the bold whole-brain mask in sessions/<session_id>/images/segmentation/boldmasks/bold[N]_frame1_brain_mask.nii.gz or the freesurfer segmentation in sessions/<session_id>/images/segmentation/freesurfer/mri/aparc+aseg_bold.nii.gz,
respectively.
Any additional extracted signal will be added as another named column in the generated .nuisance file.
Regression of BOLD nuisance signal
The extracted BOLD nuisance signal is regressed in the GLM step implemented in preprocess_bold and preprocess_conc commands similar to the motion correction parameters. To remove an extracted nuisance signal, specify it with the --bold_nuisance parameter. Any signal present in the .nuisance file can be added to the comma-separated list. E.g. --bold_nuisance="V, WM,WB,eyes,scull. If 'n1d' or '1d' is added to the list, the first derivative of all BOLD nuisance signals or all specified nuisance signals (including motion correction parameters) will also be added to the GLM regression.
Regression of additional signals
In addition to the motion correction parameters and the nuisance signals extracted from the BOLD images, any other signal can be added to the nuisance signal regression. These can include various physiological measures, such as blood oxygen levels, heart rate, respiration, pupil size,
EEG alpha power, and any other signals of potential interest, such as eye movements, external motion parameters, and others. QuNex does not natively provide functions to process and add these signals, but they can be added as additional columns to the .nuisance files manually or using custom scripts. Any column in the .nuisance files can then be referenced with the --bold_nuisance parameter, just like previously described motion correction parameters and BOLD nuisance signals.
The functionality described allows control, removal or estimation of any continuous signal in the GLM modeling step. For more information and details, please consult the following additional sources:
- inline help for
extract_nuisance_signal,preprocess_boldandpreprocess_concQuNex commands - inline help for
general_extract_nuisance,fc_preprocess_boldandfc_preprocess_concmatlab functions - file specification for
.namesROI definition files
Written by Grega Repovš
Last update: 2022-07-08
QuNex version: 0.93.8
